Venture into the Dark Forest of MS Spectra:
Where Precious (Epi)Lipids hide Themselves in Shadows.
Embark on a Journey of Unending Discovery!
# Crafting insights from omics data.
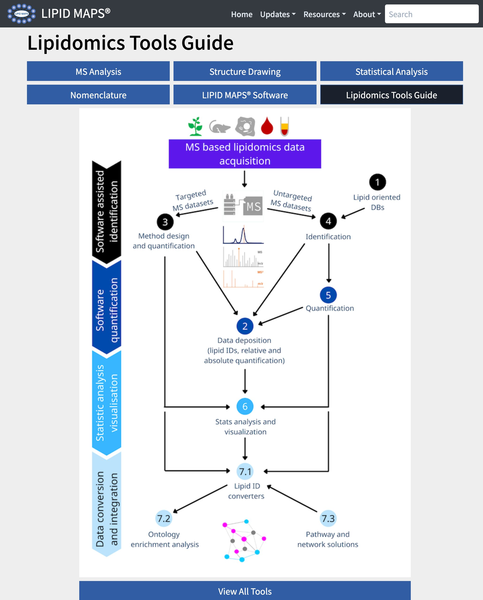
Lipidomics Tools Guide
- Lipidomics Tools Guide on LIPID MAPS® website
- Ni, Zhixu, Michele Wölk, Geoff Jukes, Karla Mendivelso Espinosa, Robert Ahrends, Lucila Aimo, Jorge Alvarez-Jarreta et al. “Guiding the choice of informatics software and tools for lipidomics research applications.” Nature methods 20, no. 2 (2023): 193-204.
Together with LIPID MAPS we designed a Lipidomics Tools Guide. Lipidomics Tools Guide summarizes multiple software tools with open access to the academic users addressing different steps in the lipidomics data processing pipeline including lipid-oriented databases, MS data repositories, analysis of targeted and untargeted lipidomics datasets, lipid quantification, statistical analysis, visualization, and data integration solutions.
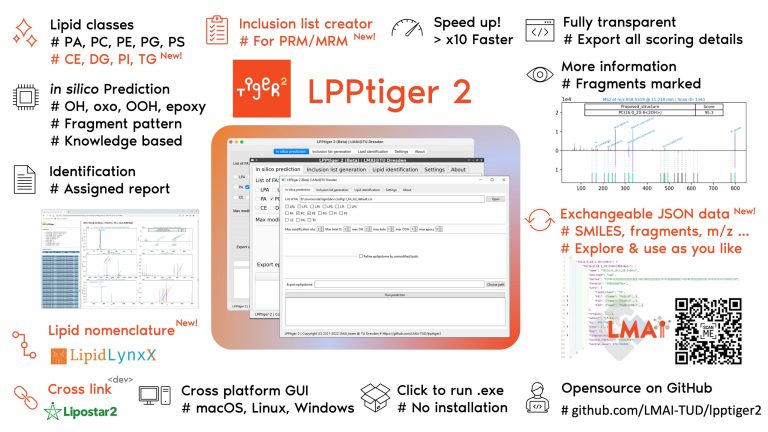
LPPtiger 2
- Download LPPtiger2 (for Windows)
- Source code on GitHub (for Linux, macOS & Windows)
- Ni, Zhixu, Georgia Angelidou, Ralf Hoffmann, and Maria Fedorova. “LPPtiger software for lipidome-specific prediction and identification of oxidized phospholipids from LC-MS datasets.” Scientific reports 7, no. 1 (2017): 15138.
LPPtiger2 is our flagship tool for analysis of oxidized complex lipids. It provides all-in-one-solution for our epilipidomics workflow, starting with knowledge based in silico prediction of modified lipids, in silico fragmentation using accurately defined rules, and identification from LC-MS/MS datasets. It also has the function to directly generate inclusion list for PRM experiments. LPPtiger2 provides interactive HTML output with its unique six-panel-image, which allows to review, store, and share the identification results.
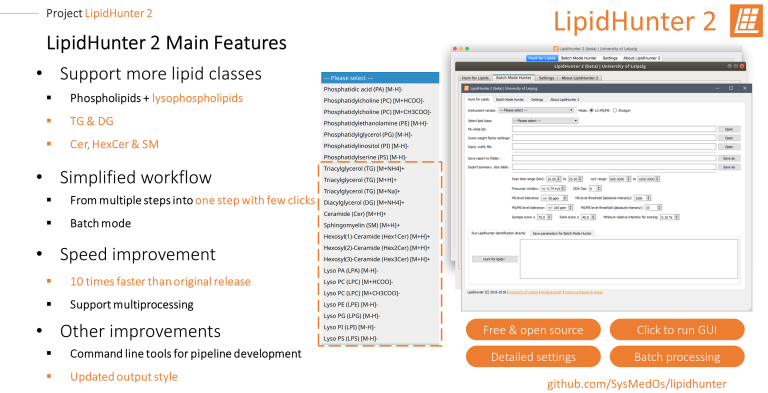
LipidHunter 2
- Download LipidHunter2 (for Windows)
- Source code on GitHub (for Linux, macOS & Windows)
- Ni, Zhixu, Georgia Angelidou, Mike Lange, Ralf Hoffmann, and Maria Fedorova. “LipidHunter identifies phospholipids by high-throughput processing of LC-MS and shotgun lipidomics datasets.” Analytical Chemistry 89, no. 17 (2017): 8800-8807.
LipidHunter2 is capable to perform bottom-up identification of lipids from LC-MS/MS and shotgun lipidomics data by resembling a workflow of manual spectra annotation. LipidHunter provides interactive HTML output with its unique six-panel-image, which allows to review, store, and share the identification results.
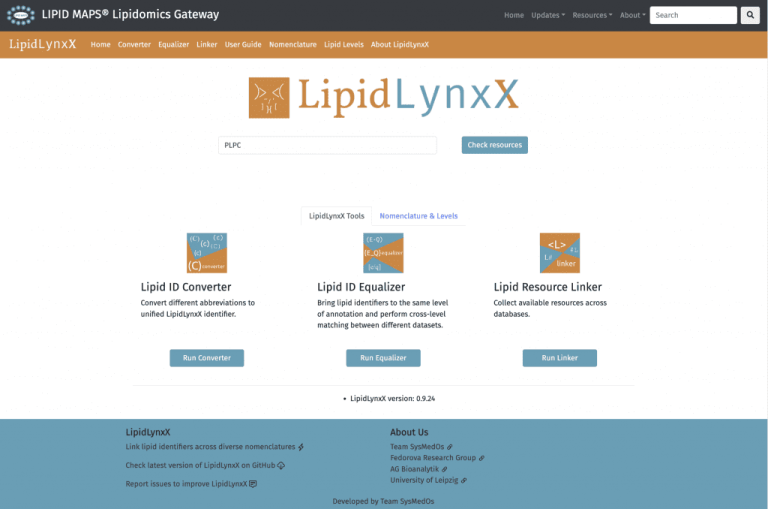
LipidLynxX
- LipidLynxX online tool powered by LIPID MAPS
- Source code on GitHub (for Linux, macOS & Windows)
- Ni, Zhixu, Michele Wölk, Geoff Jukes, Karla Mendivelso Espinosa, Robert Ahrends, Lucila Aimo, Jorge Alvarez-Jarreta et al. “Guiding the choice of informatics software and tools for lipidomics research applications.” Nature methods 20, no. 2 (2023): 193-204.
Modern high throughput lipidomics provides large-scale datasets reporting hundreds of lipid molecular species. However, cross-laboratory comparison, meta-analysis, and systems biology integration of in-house generated and published datasets remain challenging due to a high diversity of used lipid annotation systems, different levels of reported structural information, and shortage in links to data integration resources. To support lipidomics data integration and interoperability of experimental lipidomics with data integration tools, we developed LipidLynxX serving as a hub facilitating data flow from high-throughput lipidomics analysis to systems biology data integration. LipidLynxX allows conversion from 25 different annotation styles, cross-matching of lipid datasets with different levels of structural confirmation, and linking of diverse lipid annotations to six different tools supporting lipid ontology, pathway, and network analysis aiming systems-wide integration and functional annotation of lipidome dynamics in health and disease. LipidLynxX is a flexible, customizable open-access tool freely available for download at https://github.com/SysMedOs/LipidLynxX as well as via web-interface on LIPID MAPS, at http://lipidmaps.org/lipidlynxx/.